Whole Body Regeneration model: Botrylloides sp.
Animals capable of some sort of regeneration are distributed widely throughout most phyla. The most dramatic example of such the regenerative process is whole-body regeneration (WBR), the process of regenerating an entire adult organism from only a sm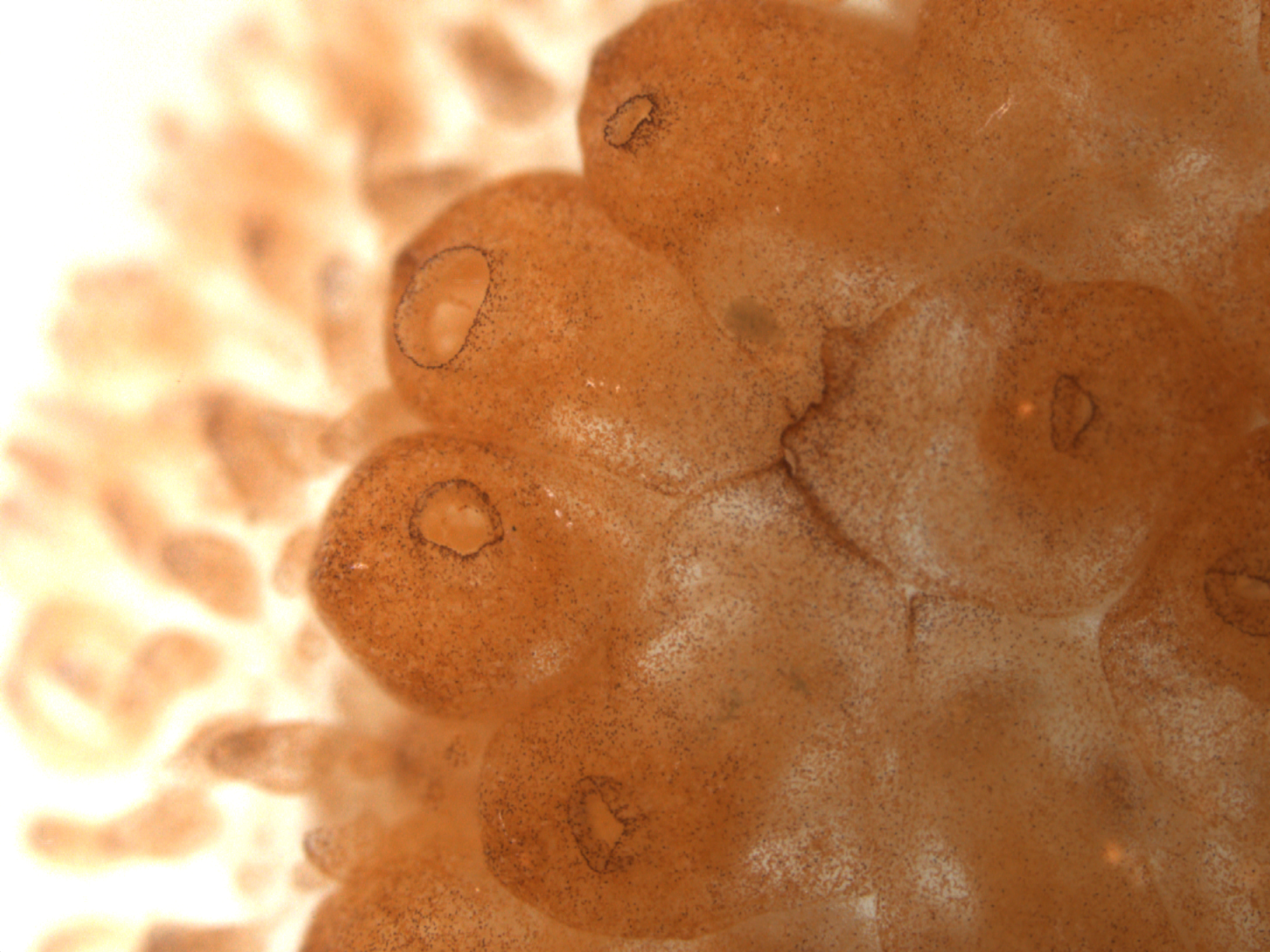 all number of cells or amount of tissue. From an evolutionary perspective, as animal body and tissue complexity increases, regenerative (and particularly WBR) ability decreases.
all number of cells or amount of tissue. From an evolutionary perspective, as animal body and tissue complexity increases, regenerative (and particularly WBR) ability decreases.
An exception to this is Botrylloides diegensis, an invertebrate colonial sea squirt that can undergo WBR. As a chordate (processing a notochord), B. leachi is a close phylogenetic relative of vertebrates and provides an excellent model for vertebrate regeneration.
Minute fragments of vascular tissue from a colony, containing only a few hundred blood cells are capable of regenerating a whole functional adult organism within 7-10 days. We have sequenced the genome, regeneration transcriptomes to take advantage of this WBR model to investigate the molecular and cellular triggers of regeneration.
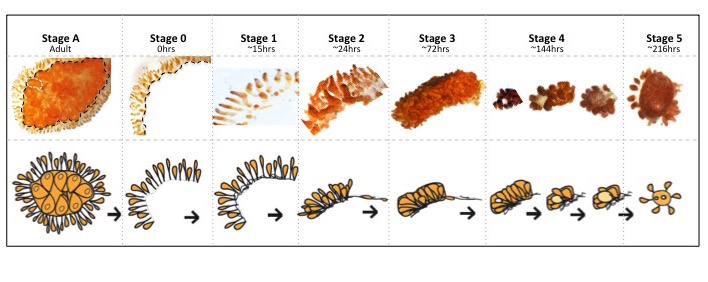 Stage A: Botrylloides colony collected from the marina at Dunedin, NZ, before any dissection had occurred. Black line indicating dissection site. Stage 0: Marginal ampullae at 0 h, directly after dissection from the zooids. Stage 1: New vascular connections formed between ampullae. Stage 2: Marginal ampullae started to condense together creating a compact network of blood vessels within the tunic matrix. Stage 3: Blood vessels more condensed. Stage 4: Formation of small transparent vesicle in the middle of the condensed blood vessels. This transparent ball continues to expand in size and eventually gains pigment before forming the new adult. Stage 5: A fully functioning zooid capable of filter feeding
Stage A: Botrylloides colony collected from the marina at Dunedin, NZ, before any dissection had occurred. Black line indicating dissection site. Stage 0: Marginal ampullae at 0 h, directly after dissection from the zooids. Stage 1: New vascular connections formed between ampullae. Stage 2: Marginal ampullae started to condense together creating a compact network of blood vessels within the tunic matrix. Stage 3: Blood vessels more condensed. Stage 4: Formation of small transparent vesicle in the middle of the condensed blood vessels. This transparent ball continues to expand in size and eventually gains pigment before forming the new adult. Stage 5: A fully functioning zooid capable of filter feeding
To find out more – Whole Body Regeneration model: Botrylloides leachi
Team: Michael Meier, Rebecca Clarke, Beri Temiz
The evolution of ancient developmental pathways – from invertebrates to vertebrates.
How do such critical pathways evolve? We study the changes that occur to gene regulatory sequences and associated transcriptional factor families and how this impacts on the evolution of developmental processes. How these families take on new roles during evolution or divide up their ancestral functions following gene duplication? For this we are making use of three models, the chordate Ciona savignyi and the vertebrates, mouse and zebrafish. We focus on the SOX family of transcription factors. These are DNA-binding proteins that regulate a diverse range of developmental processes including organogenesis and cell fate determination. Sox genes are found in the genomes of all animals and have expanded in the vertebrate lineages.